rrcf
🌲 Implementation of the Robust Random Cut Forest Algorithm for anomaly detection on streams
View the Project on GitHub kLabUM/rrcf
Theory
• Related work and motivation• Tree construction
• Insertion and deletion of points
• Anomaly scoring
Basics
• RCTree data structure• Modifying the RCTree
• Measuring anomalies
• API documentation
• Caveats and gotchas
Examples
• Batch detection• Streaming detection
• Analyzing taxi data
• Classification
• Comparison of methods
Batch anomaly detection
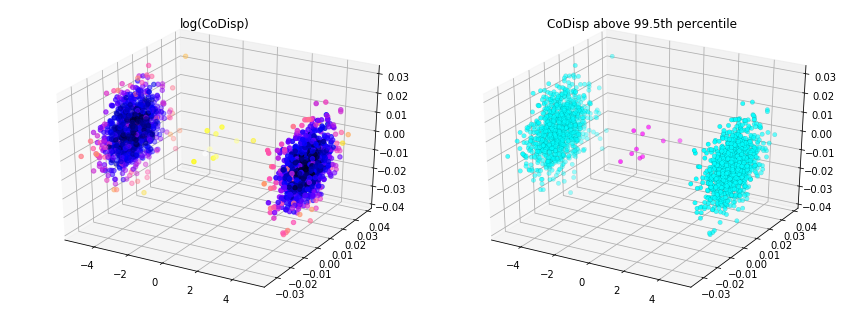
This example shows how a robust random cut forest can be used to detect outliers in a batch setting. Outliers correspond to large CoDisp.
Import modules and create sample data
import numpy as np
import pandas as pd
import rrcf
# Set sample parameters
np.random.seed(0)
n = 2010
d = 3
# Generate data
X = np.zeros((n, d))
X[:1000,0] = 5
X[1000:2000,0] = -5
X += 0.01*np.random.randn(*X.shape)
Construct a random forest
# Set forest parameters
num_trees = 100
tree_size = 256
sample_size_range = (n // tree_size, tree_size)
# Construct forest
forest = []
while len(forest) < num_trees:
# Select random subsets of points uniformly
ixs = np.random.choice(n, size=sample_size_range,
replace=False)
# Add sampled trees to forest
trees = [rrcf.RCTree(X[ix], index_labels=ix)
for ix in ixs]
forest.extend(trees)
Compute anomaly score
# Compute average CoDisp
avg_codisp = pd.Series(0.0, index=np.arange(n))
index = np.zeros(n)
for tree in forest:
codisp = pd.Series({leaf : tree.codisp(leaf)
for leaf in tree.leaves})
avg_codisp[codisp.index] += codisp
np.add.at(index, codisp.index.values, 1)
avg_codisp /= index
Plot result
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.pyplot as plt
from matplotlib import colors
threshold = avg_codisp.nlargest(n=10).min()
fig = plt.figure(figsize=(12,4.5))
ax = fig.add_subplot(121, projection='3d')
sc = ax.scatter(X[:,0], X[:,1], X[:,2],
c=np.log(avg_codisp.sort_index().values),
cmap='gnuplot2')
plt.title('log(CoDisp)')
ax = fig.add_subplot(122, projection='3d')
sc = ax.scatter(X[:,0], X[:,1], X[:,2],
linewidths=0.1, edgecolors='k',
c=(avg_codisp >= threshold).astype(float),
cmap='cool')
plt.title('CoDisp above 99.5th percentile')